"It takes all the running you can do, to keep in the same place."
- The Red Queen to Alice, in Lewis Carroll’s Through the Looking Glass
Pathogens and their hosts are locked in a constant struggle for evolutionary dominance. Any host adaptation that leads to more effective defense against a pathogen will almost inevitably drive adaptation in the pathogen to escape such defenses. Thus, the only way either side can survive is to continually adapt, as proposed in Leigh Van Valen’s ‘Red Queen hypothesis’. These escalating 'arms races' play out at the level of rapid evolution of host-pathogen interactions, constantly selecting for molecular innovations that either protect the host or evade host defenses. However, the molecular details of these rapidly evolving host-pathogen interactions, and the consequences of these conflicts on human susceptibility to current and emerging infectious diseases, are mostly unexplored.
Our research aims to use these evolutionary principles, combined with virology, immunology, and mechanistic biochemistry, to understand how the immune system has evolved to defend against pathogens and how pathogens counter-evolve to defeat host immunity. One of the approaches we take is to use comparative genomics to identify characteristic signatures of pathogen-driven innovation (e.g. rapid sequence evolution and/or extensive human polymorphism, horizontal gene transfer, gene birth and death, gene recombination). Interestingly, we expect these rapidly evolving genes to be at the center of host-pathogen conflicts, yet many of them are almost entirely uncharacterized. This provides an opportunity to use pathogens to discover novel biological functions that are involved in not only pathogen immunity but also more global cellular regulation. The ultimate goal of our research is to understand the rapidly evolving molecular means by which hosts and pathogens battle each other, while at the same time providing insight into how genetic variation resulting from these conflicts contributes to differences in human susceptibility to infectious diseases, including zoonotic transmission of viruses from other species.
Current projects:
Distiguishing 'self' from 'non-self' mRNA during viral infection
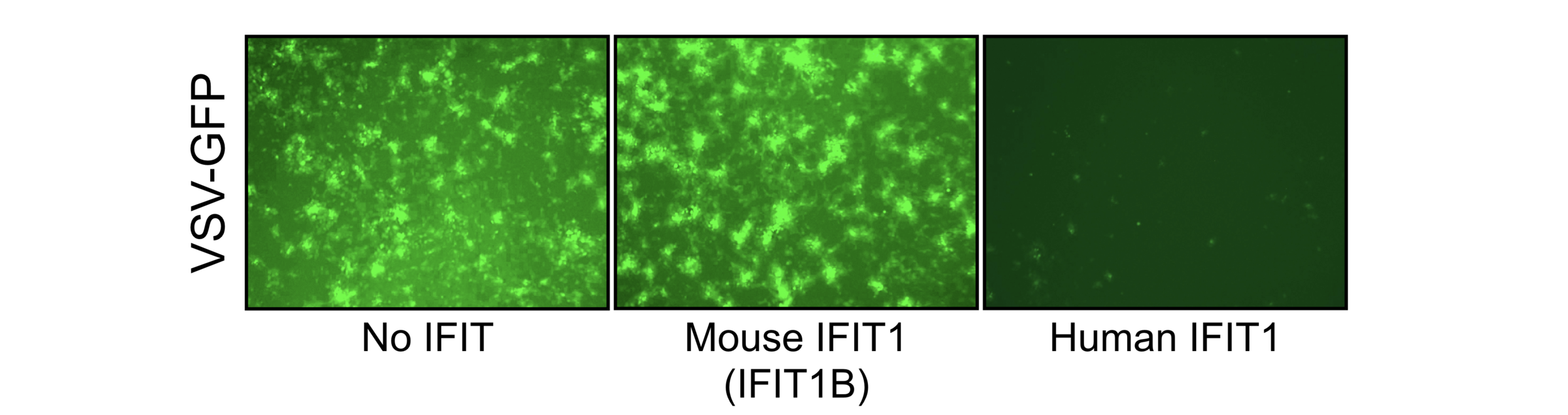
A common mechanism in antiviral immunity is to distinguish between self (e.g. host) mRNA and non-self (e.g. pathogen) mRNA. We are interested in understanding how post-transcriptional modifications of mRNA, or other mRNA features, serve as such 'self versus non-self' patterns in the innate antiviral response. Our evolutionary analyses highlight the involvement of several IFIT (interferon-induced with tetratricopeptide repeats) proteins as important, and rapidly evolving, sensors of different mRNA modifications. Likewise, several host and viral RNA modification enzymes are highly evolutionarily dynamic. Based on these data, and the widespread turnover of IFIT genes across mammals, we expect that rapidly evolving mRNA modification and recognition systems contribute to differences in antiviral specificity between different species.
Post-translational modifications at the interface of host-virus conflicts
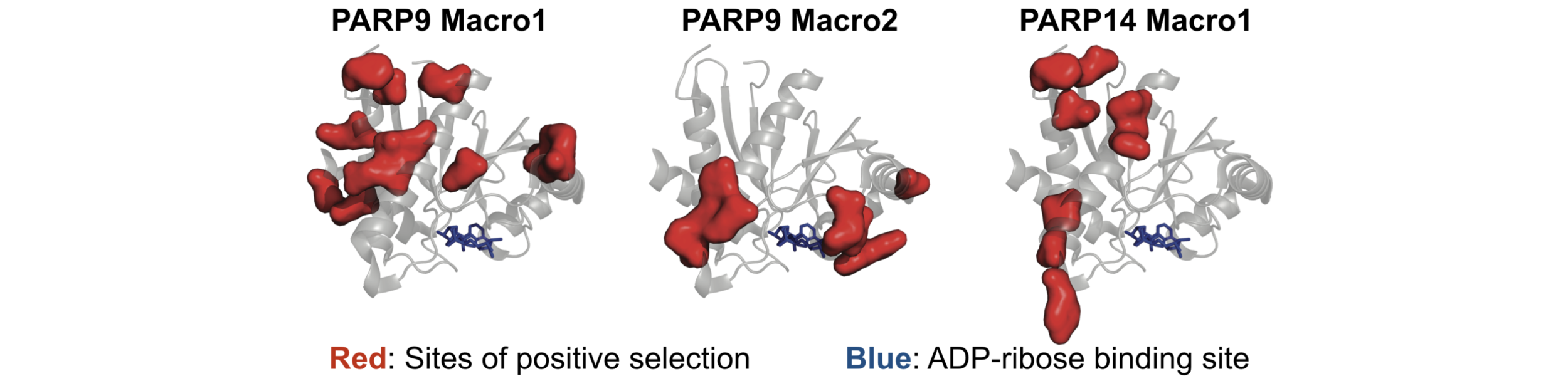
It is increasingly obvious that post-translational modification of proteins is a common battleground for host-pathogen conflicts. We are using evolutionary signatures of innovation in genes involved in adding, recognizing and removing ('writers', 'readers' and 'erasers') post-translational modifications to characterize a molecular role for them in host-pathogen interactions. One particular focus is on ADP-ribosylation by members of the PARP (poly ADP-ribose polymerase) gene family, as several uncharacterized members of this gene family are induced by the innate immune system and are very rapidly evolving. By determining how this modification is being utilized by the immune system, and how it is being subverted or antagonized by viruses, we hope to gain greater insight into how this poorly understood protein modification is generally used to regulate cellular processes.
Viral proteases as drivers of host gene evolution and determinants of host range
In addition to discovering novel mechanisms of antiviral immunity, we are also interested in discovering the viral factors that are interacting with these host proteins. Our current focus is on proteases that are encoded by many human viruses, including picornaviruses (e.g. human rhinovirus), flaviviruses (e.g. dengue virus), coronaviruses (e.g. SARS-CoV-2), alphaviruses (e.g. Chikungunya virus) and more. These proteases cleave host proteins at sequence-specific locations, setting up an immediate potential for back-and-forth evolutionary arms races between proteases and these host proteins. We are using this as a way to understand virus-specific and host-specific determinants of infection, including how human polymorphism shapes differences in the antiviral immune response. Within this project, we are particularly interested in discovering and characterizing 'tripwire' sensors of viral proteases, such as human NLRP1, which are activated rather than inhibited upon viral cleavage.
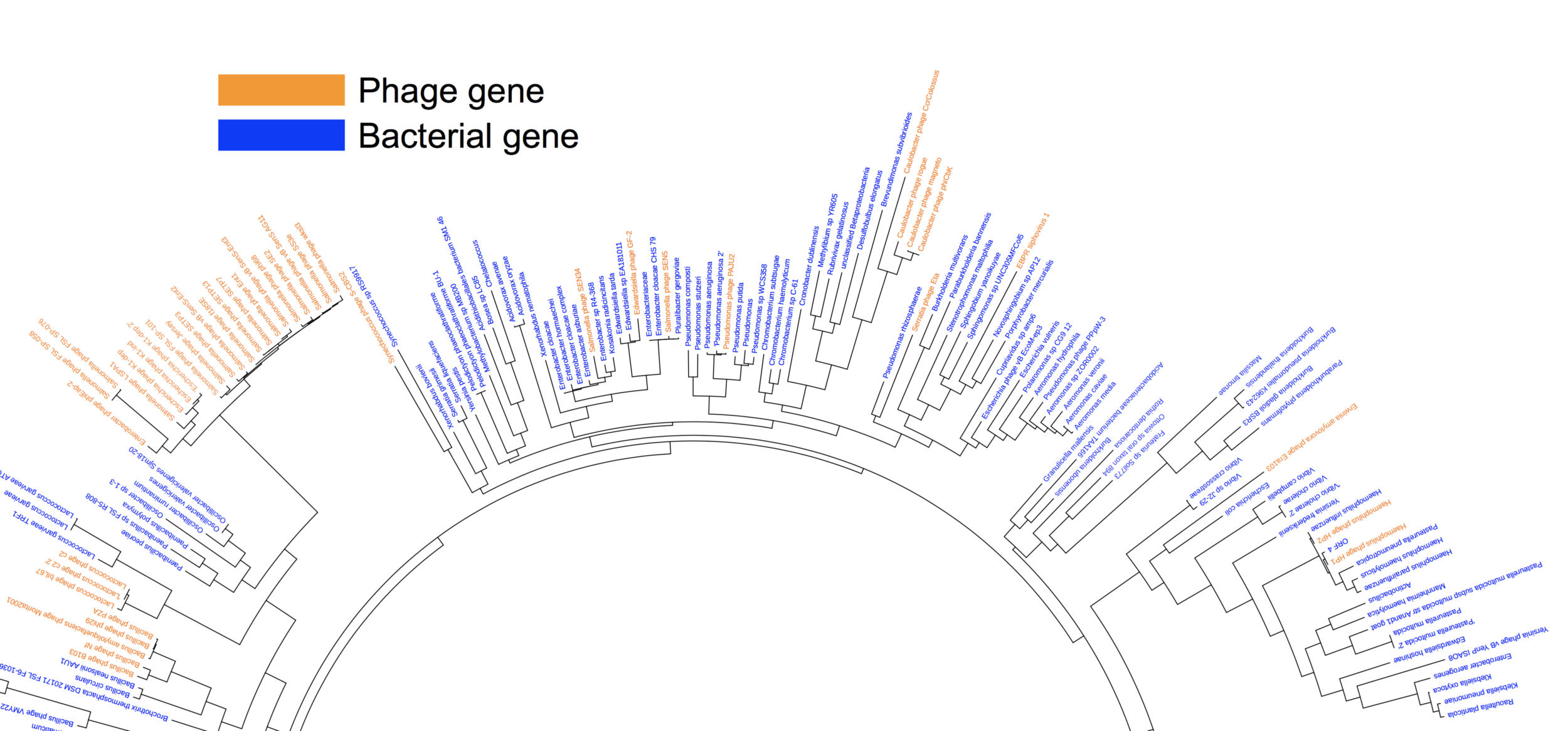
Horizontal gene transfer as a source of genetic innovation for hosts and their pathogens
Genetic innovations that are employed during host-pathogen arms races can come in many different flavors. One of the most striking forms of innovation comes by the movement of genetic material from one organism into another by horizontal gene transfer (HGT). Such a mechanism allows organisms to essentially steal genes from one another and then deploy these stolen genes in ongoing host-pathogen conflicts. To understand the role of HGT in host-pathogen evolution, we are developing methods to rapidly identify instances of HGT into metazoans (multicellular animals). In addition to identifying these rare events, we are interested in characterizing the functional consequences of HGT, especially in terms of novel immune functions that have evolved from 'stolen' microbial genes.